Metagenomic sequencing is a culture-independent, high-throughput analysis to identify microbial composition (e.g. bacteria, virus, and fungi) in food, soil, skin and other environments. Taxonomic analyses and functional profiling make metagenomics a powerful tool to understand the role of microorganisms in a complex community. IMGM metagenomic services are DAkkS accredited according to ISO17025.
Using MiSeq and NovaSeq sequencing technologies, IMGM offers the following services:
Services | Target | Purpose | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 16S Amplicon-Seq | V1-2, V3-V4* or V6-V8 region of 16S rRNA genes | prokaryotic identification | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 18S Amplicon-Seq | V1-V3 region of 18S rRNA genes | eukaryotic identification | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| ITS Amplicon-Seq | ITS1, ITS2 region or 5.8S rDNA | fungal identification | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
weitere Inhalte anzeigen | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
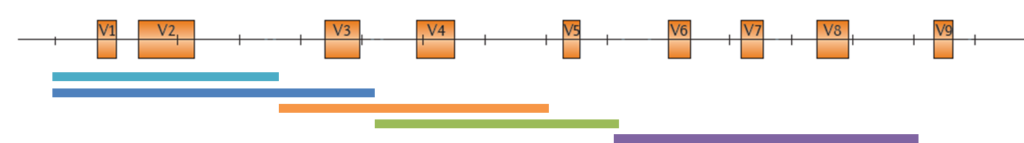
Focusing on specific marker genes for phylogenetic identification/classification is one of the main categories of metagenomic sequencing. For this so-called targeted amplicon approach, the ribosomal small subunit RNA (16S rRNA) gene has emerged as the most commonly used marker. Further target genes may include other rRNA genes, the ITS region, COI, Rubisco or additional functional genes.
IMGM’s flexible metagenomics amplicon sequencing service offers the choice between experimentally tested primer pairs for different variable regions alongside the 16S rRNA gene (see graph below).
We also offer you a completely flexible choice for own primers depending on your targets and research aims. Our project managers will support you from custom Primer design to individualized bioinformatics.
Additional flexibility is provided by the possibility to step into the analysis pipeline at your most convenient starting point, e.g. start directly with gDNA, preamplified PCR products or even sequencing-ready libraries. If your project belongs to the category “off the beaten path”, IMGM has a long-standing expertise with scientific consultation and custom project design for metagenomics analyses.
For 16S/18S and ITS we offer the analysis of microbial composition, diversity and abundance with up-to-date bioinformatics software tools. OTU clustering and phylogenetic classification as well as further steps like calculation of alpha and beta diversity is carried out with a diligent view on data quality. This enables you to easily extract conclusions from your experimental data and to directly use our data to publish your results.
Use of the entire DNA for sequencing is often referred to as Shotgun Metagenomics or, in case of RNA, as Metatranscriptomics. These approaches offer a random representation of all extracted genomic sequences, giving insights into the metabolic profiles of these specific communities.
Unlike the targeted approach used in amplicon sequencing, Shotgun Metagenomic Sequencing is performed to provide not only information on the taxonomic annotations of each organism, but also on the whole genetic content of your sample. Have a look at its gene expression pattern, find potentially novel biocatalysts or enzymes or get evolutionary profiles of community function and structure.
- Selection between IMGM´s standard portfolio and custom primers
- Flexible starting point from DNA isolation to sequencing-ready library loading
- Professional consulting and proposal of the appropriate technical approach and platform
- Reliable sample processing throughout the workflow
- Fast turnaround times and focus on high-quality data
- Customized bioinformatic panels
- Detailed and comprehensive reporting of data and results
- Dedicated service for medical/clinical samples